How DNA could solve the world’s data storage problem

Data overload is a uniquely 21st century problem. Could the answer be as old as life itself?
The amount of data generated in today’s digital era is truly astounding. In the last two years alone more data was generated than in the previous millennia, and our hunger for data is showing no signs of letting up.
Our current infrastructure and hardware will soon be unable to accommodate the incoming deluge of information, which is otherwise expected to consume all microchip grade silicon by 2040. To address this uniquely modern problem, industries are looking to an ancient alternative: DNA.
The potential of DNA as a storage medium has long been touted by scientists: its compactness, ease of replication and longevity seen as the clear advantages. This shouldn’t come as a shock, remembering that DNA has literally been the storage medium for all life on earth for probably as long as it has existed, however our ability to manipulate DNA in such a way to take advantage of these features is a far more recent discovery.
Advances over the last few decades in our ability to synthesise (write) and sequence (read) DNA have meant that this concept is now a reality; all kinds of media including Shakespeare’s sonnets, Deep Purple’s “Smoke on the Water” and a GIF of a galloping horse have all been successfully stored and retrieved from the seemingly invisible DNA strands. So how does this all work?
DNA encodes information through its constituent parts. Each DNA molecule is a double-stranded polymer composed of four nucleotides: A (adenine) and T (thymine) structurally referred to as purines; C (cytosine) and G (guanine), the pyrimidines. In nature, three of these bases —known as a codon— encodes for a single amino acid, which in turn form the proteins which constitute life.
In more data-minded terms, being one of four possible nucleotides, each nucleotide represents a maximum of two bits of information. For reference: eight bits make up a byte; one thousand (well, 1024 to be precise) bytes make a kilobyte; another thousand kilobytes is a megabyte; and a thousand megabytes is a gigabyte. A 300 page book corresponds to around 400 kilobytes, an average .mp3 track is around 4 megabytes, and HD films typically run to 5 gigabytes. Because DNA is so compact, a single gram of it has a theoretical capacity of 215 million gigabytes, which corresponds to about 600 trillion books. This means that if stored as DNA, all of humanity’s current data could be contained within a single room.
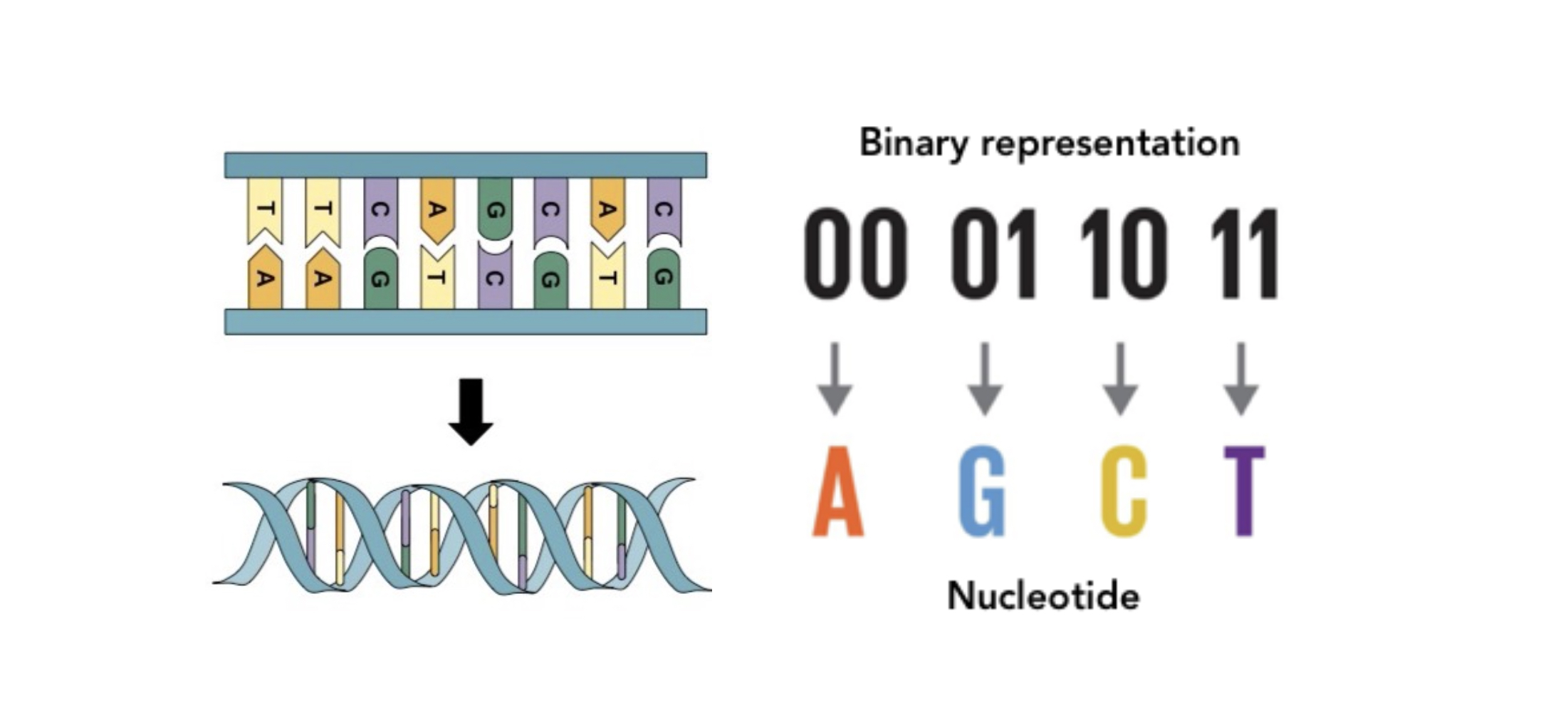
DNA storage. The structure of DNA, showing the four nucleotides (left) , and how they could encode binary information (right).
DNA may also alleviate another issue plaguing data storage: hardware obsolescence. While storage devices and associated hardware tend to become obsolete over time —think floppy discs and CDs— it is unlikely that we will ever lose interest in sequencing DNA. It also won’t degrade as quickly as magnetic tape or CDs - the devices currently being used to house most digital archives - especially if stored under optimal conditions.
The main problem, of course, is cost. Although synthesising and sequencing DNA has become far less expensive in recent years, it still represents the biggest barrier to commercial use of the technology. For example, when Yaniv Erlich and Dina Zielinski of Columbia Univerity and the New York Genome Centre stored and retrieved a number of files using DNA in 2017, it cost them approximately $5000 USD per megabyte.
Dina Zielinski, a bioinformatics expert, explains why DNA storage is needed and how it could work. Video: TED.com
While these costs are showing exponential decreases over time, some organisations are taking a more direct route to reducing costs. DNA storage start-up Catalog is rethinking the entire process by decoupling the process of synthesising DNA and encoding data.
In traditional methods, the information is encoded, through 1s and 0s, as a sequence of bases in each strand. Instead, Catalog’s idea is to use a relatively small number of prefabricated DNA fragments, each no longer 30 bases, which can then be combined in an almost infinite number of ways to encode information by using enzymatic reactions. This would reduce the amount of DNA synthesis required: the most expensive and time-consuming part.
Catalog hope that this approach should enable the company to achieve costs competitive with other storage devices in a matter of years. Still, there are a number of other engineering challenges which must be solved before this technology is adopted commercially.
In the short term, it is more likely that we’ll see DNA being used for rarely-accessed archives, or by industries such as film studios, research laboratories or government agencies which need to store large amounts of data indefinitely. Given more time, however, DNA may become a key part of our infrastructure, storing not only the blueprint for life but all of the data that comes along with it.